The CRISPR gene editing technology that was born in recent years has completely changed molecular biology and given us unprecedented ability to precisely modify the genome. However, conventional CRISPR gene editing technology produces DNA double-strand breaks (DSBs), which may cause adverse events or even serious fatal risks in clinical treatment, including unwanted insertion or deletion mutations, chromosome-scale mutations, p53 activation, and cell cycle arrest. In addition, gene editing technology that relies on homology-directed repair (HDR) depends on the cell division cycle and is therefore not applicable to all cell types.
The next generation of gene editing technology attempts to overcome the above two shortcomings. Among them, base editing and prime editing have proven to be successful, but they also have shortcomings such as limited editing range, low editing efficiency, and insufficient ease of use.
On July 22, 2024, Benjamin P. Kleinstiver's team from Harvard Medical School and Massachusetts General Hospital published a research paper titled "Click editing enables programmable genome writing using DNA polymerases and HUH endonucleases" in Nature Biotechnology.
This study developed a new gene editing technology called click editing (CE), which combines HUH endonuclease (HUHe), DNA-dependent polymerase (DDP) and RNA-guided Cas9 nickase (nCas9), and can perform programmable precise genome engineering from simple DNA templates, including replacement, insertion and deletion of the genome. This research result points out a new direction for the development of next-generation gene editing technology.
Recently, studies have developed gene editing technologies that can change DNA sequences without generating DSBs, such as base editing and prime editing developed by Ruqian Liu's team. However, base editing systems are prone to unnecessary off-target editing and can only edit a single base, while prime editing systems have disadvantages such as low editing efficiency and complex pegRNA design.
Compared with other types of gene editors, the DDP-based genome editing technology developed in this study shows unparalleled potential. DDP is ubiquitous in cells, may have enzymatic activity in almost any type of cell, and has advantages such as high affinity and high-fidelity polymerization. In addition, DDP is compatible with widely used, clinically validated and user-specifiable DNA oligonucleotide templates, which may have advantages in simplicity, scalability, customizability, chemical stability and editing purity.
In this latest study, the research team envisioned that fusing or recruiting DDP to a DNA nickase (nCas9) might create a class of gene editing technologies that differ from previous methods. The research team speculated that the use of a single-stranded DNA (ssDNA) recruitment domain could enable the localization of the modified coding template, thereby improving editing efficiency.
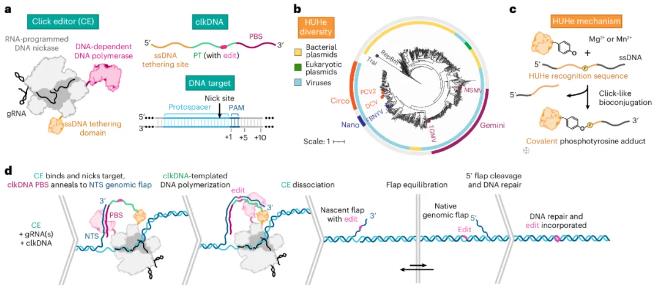
Figure 1. Overview and development of click editing. (Ferreira da Silva, et al., 2024)
The ssDNA template should consist of three parts: a recognition sequence that binds to the nuclease protein or polypeptide; a polymer template (PT) that achieves editing replacement; and a primer binding site (PBS) with homology to the non-target strand (NTS) of the target site.
The ideal ssDNA recruitment domain should be specific to the ssDNA template, have a small coding sequence, catalyze the dynamics of the fusion protein-DNA complex, and not require any specialized modifications. HUHe are a family of proteins that meet these criteria. HUHe is a class of small proteins that are distributed in most species and perform various ssDNA-specific activities, including ssDNA viral replication, conjugation, and transposition.
Based on these ideas, the research team developed click editors, in which nCas9 can target the target genomic site and produce a cut under the guidance of guide RNA (gRNA), HUHe covalently connects the click DNA (clkDNA) template encoding the desired edit to the genomic DNA at the cut site, and then DPP uses these templates to accurately introduce gene editing.
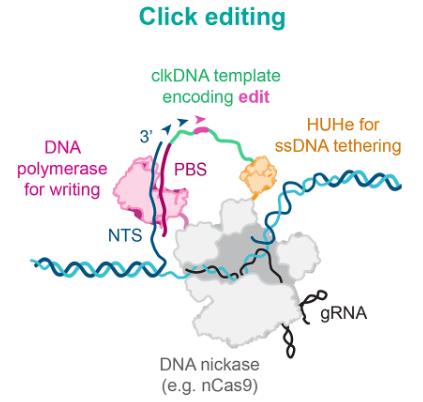
Figure 2. Structural components of click editors. (Ferreira da Silva, et al., 2024)
Not only that, the simplicity of using unmodified DNA molecules as clkDNA templates allows for rapid optimization of editing efficiency through different parameters, including editing type and additional silent mutations. In addition, clkDNA can be screened in high throughput without the need for cumbersome molecular cloning steps.
The click editing system is modular in composition and can use various DDP or HUHe domains, with the potential for further optimization and engineering. Through iterative optimization of the modular components of click editing and its clkDNA, the research team demonstrated the ability to perform precise genome editing with minimal insertion or deletion mutations in a variety of immortalized human cell types and primary fibroblasts, and the precise editing efficiency is as high as 30%.
More importantly, the click editing system can achieve multiple types of editing, including base substitution, short DNA fragment insertion or deletion, and excels in programmability, versatility and precision, and does not produce DSBs, does not rely on HDR, and can be applied to various biological applications.
Overall, this study developed a new gene editing technology - click editing, which uses HUHe-mediated covalent clkDNA to locate the target site of DDP, thereby achieving precise and universal genome writing. This result highlights the advantages of DDP in gene editing technology. The modularity and versatility of click-editing systems allow them to be applied to a wide range of species and continue to be developed and optimized as a general tool for a variety of applications, thus opening up new research directions in the field of gene editing.
| Cat# | Product Name | Size |
| ACC-100 | GV3101 Chemically Competent Cell | 10 tubes (100μL/tube) 20 tubes (100μL/tube) 50 tubes (100μL/tube) 100 tubes (100μL/tube) |
| ACC-103 | EHA105 Chemically Competent Cell | 10 tubes (100μL/tube) 20 tubes (100μL/tube) 50 tubes (100μL/tube) 100 tubes (100μL/tube) |
| ACC-105 | AGL1 Chemically Competent Cell | 10 tubes (100μL/tube) 20 tubes (100μL/tube) 50 tubes (100μL/tube) 100 tubes (100μL/tube) |
| ACC-107 | LBA4404 Chemically Competent Cell | 10 tubes (100μL/tube) 20 tubes (100μL/tube) 50 tubes (100μL/tube) 100 tubes (100μL/tube) |
| ACC-108 | EHA101 Chemically Competent Cell | 10 tubes (100μL/tube) 20 tubes (100μL/tube) 50 tubes (100μL/tube) 100 tubes (100μL/tube) |
| ACC-117 | Ar.Qual Chemically Competent Cell | 10 tubes (100μL/tube) 20 tubes (100μL/tube) 50 tubes (100μL/tube) 100 tubes (100μL/tube) |
| ACC-118 | MSU440 Chemically Competent Cell | 10 tubes (100μL/tube) 20 tubes (100μL/tube) 50 tubes (100μL/tube) 100 tubes (100μL/tube) |
| ACC-119 | C58C1 Chemically Competent Cell | 10 tubes (100μL/tube) 20 tubes (100μL/tube) 50 tubes (100μL/tube) 100 tubes (100μL/tube) |
| ACC-121 | K599 Chemically Competent Cell | 10 tubes (100μL/tube) 20 tubes (100μL/tube) 50 tubes (100μL/tube) 100 tubes (100μL/tube) |
| ACC-122 | Ar.A4 Electroporation Competent Cell | 10 tubes (50μL/tube) 20 tubes (50μL/tube) 50 tubes (50μL/tube) |